The SciSpacy project from AllenAI provides a language model trained on biomedical text, which can be used for Named Entity Recognition (NER) of biomedical entities using the standard SpaCy API. Unlike the entities found using SpaCy's language models (at least the English one), where entities have types such as PER, GEO, ORG, etc., SciSpacy entities have the single type ENTITY. In order to further classify them, SciSpacy provides Entity Linking (NEL) functionality through its integration with various ontology providers, such as the Unified Medical Language System (UMLS), Medical Subject Headings (MeSH), RxNorm, Gene Ontology (GO), and Human Phenotype Ontology (HPO).
The NER and NEL processes are decoupled. The NER process finds candidate entity spans, and these spans are matched against the respective ontologies, which may result in the span matching zero or more ontology entries. All candidate span is then matched to all the matched entities.
I tried annotating the
COVID-19 Open Research Dataset (CORD-19) against UMLS using the SciSpacy integration described above, and I noticed significant ambiguity in the linking results. Specifically, annotating approximately 22 million sentences in the CORD-19 dataset results in 113 million candidate entity spans, which get linked to 166 million UMLS concepts, i.e., on average, each candidate span resolves to 1.5 UMLS concepts. However, the distribution is Zipfian, with approximately 46.87% entity spans resolving to a single concept, with a long tail of entity spans being linked to up to 67 UMLS concepts.
In this post, I will describe a strategy to disambiguate the linked entities. Based on limited testing, this chooses the correct concept about 73% of the time.
The strategy is based on the intuition that an ambiguously linked entity span is more likely to resolve to a concept that is closely related to concepts for the other non-ambiguously linked entity spans in the sentence. In other words, the best target label to choose for an ambiguous entity is the one that is semantically closest to the labels of other entities in the sentence. Or even more succintly, and with apologies to John Firth, an entity is known by the company it keeps.
The NER and NEL processes provided by the SciSpacy library allows us to reduce a sentence to a collection of entity spans, each of which map to zero or more UMLS concepts. Each UMLS concept maps to one or more
Semantic Types, which represent high level subject categories. So essentially, a sentence can be reduced to a graph of semantic type using the following steps.
Consider the sentence below, the NER step identifies candidate spans that are indicated by highlights.
The fact that viral antigens could not be demonstrated with the used staining is not the result of antibodies present in the cat that already bound to these antigens and hinder binding of other antibodies.
The NEL step will attempt to match these spans against the UMLS ontology. Results for the matching are shown below. As noted earlier, each UMLS concept maps to one or more sematic types, and these are shown here as well.
| Entity-ID |
Entity Span |
Concept-ID |
Concept Primary Name |
Semantic Type Code |
Semantic Type Name |
| 1 |
staining |
C0487602 |
Staining method |
T059 |
Laboratory Procedure |
| 2 |
antibodies |
C0003241 |
Antibodies |
T116 |
Amino Acid, Peptide, or Protein |
|
|
|
|
T129 |
Immunologic Factor |
| 3 |
cat |
C0007450 |
Felis catus |
T015 |
Mammal |
|
|
C0008169 |
Chloramphenicol O-Acetyltransferase |
T116 |
Amino Acid, Peptide, or Protein |
|
|
|
|
T126 |
Enzyme |
|
|
C0325089 |
Family Felidae |
T015 |
Mammal |
|
|
C1366498 |
Chloramphenicol Acetyl Transferase Gene |
T028 |
Gene or Genome |
| 4 |
antigens |
C0003320 |
Antigens |
T129 |
Immunologic Factor |
| 5 |
binding |
C1145667 |
Binding action |
T052 |
Activity |
|
|
C1167622 |
Binding (Molecular Function) |
T044 |
Molecular Function |
| 6 |
antibodies |
C0003241 |
Antibodies |
T116 |
Amino Acid, Peptide, or Protein |
|
|
|
|
T129 |
Immunologic Factor |
The sequence of entity spans, each mapped to one or more semantic type codes can be represented by a graph of semantic type nodes as shown below. Here, each vertical grouping corresponds to an entity position. The BOS node is a special node representing the beginning of the sequence. Based on our intuition above, entity disambiguation is now just a matter of finding the most likely path through the graph.
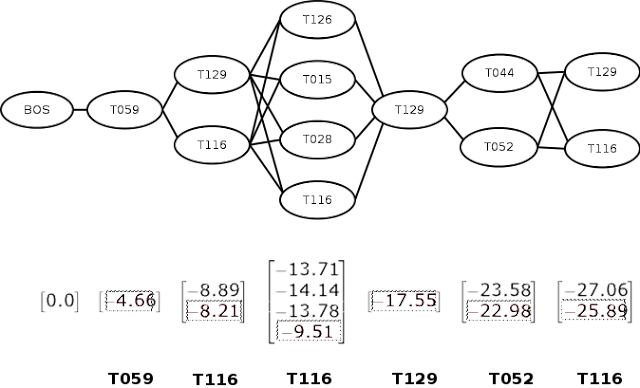
Of course, "most likely" implies that we need to know the probabilities for transitioning between semantic types. We can think of the graph as a
Markov Chain, and consider the probability of each node in the graph as being determined only by its previous node. Fortunately, this information is already available as a result of the NER + NEL process for the entire CORD-19 dataset, where approximately half of the entity spans mapped unambiguously to a single UMLS concept. Most concepts map to a single semantic type, but in cases where they map to multiple, we consider them as separate records. We compute pairwise transition probabilities across semantic types for these unambiguously linked pairs across the CORD-19 dataset and create our transition matrix. In addition, we also create a matrix of emission probabilities that identify the probabilities of resolving to a concept given a semantic type.
Using the transition probabilities, we can traverse each path in the graph from starting to ending position, computing the path probability as the product of transition probabilities (or for computational reasons, the sum of log-probabilities) of the edges. However, better methods exist, such as the
Viterbi algorithm, which allows us to save on repeated computation of common edge sequences across multiple paths. This is what we used to compute the most likely path through our semantic type graph.
The Viterbi algorithm consists of two phases -- forward and backward. In the forward phase, we move left to right, computing the log-probability of each transition at each step, as shown by the vectors below each position in the figure. When computing the transition from multiple nodes to a single node (such as the one from [T129, T116] to [T126], we compute for both paths and choose the maximum value.
In the backward phase, we move from right to left, choosing the maximum probability node at each step. This is shown in the figure as boxed entries. We can then lookup the appropriate semantic type and return the most likely sequence of semantic types (shown in bold in the bottom of the figure).
However, our objective is to return disambiguated concept linkages for entities. Given a disambiguated semantic type and multiple possibilities indicated by SciSpacy's linking process, we use the emission probabilities to choose the most likely concept to apply at the position. The result for our example is shown in the table below.
| Entity-ID |
Entity Span |
Concept-ID |
Concept Primary Name |
Semantic Type Code |
Semantic Type Name |
Correct? |
| 1 |
staining |
C0487602 |
Staining method |
T059 |
Laboratory Procedure |
N/A* |
| 2 |
antibodies |
C0003241 |
Antibodies |
T116 |
Amino Acid, Peptide, or Protein |
Yes |
| 3 |
cat |
C0008169 |
Chloramphenicol O-Acetyltransferase |
T116 |
Amino Acid, Peptide, or Protein |
No |
| 4 |
antigens |
C0003320 |
Antigens |
T129 |
Immunologic Factor |
N/A* |
| 5 |
binding |
C1145667 |
Binding action |
T052 |
Activity |
Yes |
| 6 |
antibodies |
C0003241 |
Antibodies |
T116 |
Amino Acid, Peptide, or Protein |
Yes |
(N/A: non-ambiguous mappings)
I thought this might be an interesting technique to share, hence writing about it. In addition, in the spirit of reproducibility, I have also provided the following artifacts for your convenience.
- Code: This github gist contains code that illustrates NER + NEL on an input sentence using SciSpacy and its UMLS integration, and then applies my adaptation of the Viterbi method (as described in this post) to disambiguate ambiguous entity linkages.
- Data: I have also provided the transition and emission matrices, and their associated lookup tables, for convenience, as these can be time consuming to generate from scratch from the CORD-19 dataset.
As always, I appreciate your feedback. Please let me know if you find flaws with my approach, and/or you know of a better approach for entity disambiguation



10 comments (moderated to prevent spam):
as always, very cool thinking and communication.
This is a really great approach. I will see if I manage to apply and expand it.
Thanks Alex and Matt, looking forward to what you do with it. Although I do want to point out something I learned about shortly after. The reason I built this was to figure out the best entity to match to a text span, when it matches against multiple entities in the ontology. Since I wrote this, I learned about SciSpaCy's support for EntityLinkers, which are trained models that take in a span of text identified by the Language Model, and return a list of scored of matches to different entities in its ontology. Because matches are scored, it is easy to find the best match. In addition, the EntityLinker also identifies partial matches, so it makes sense to use the EntityLinker functionality instead of this approach.
Thanks for sharing, Sujit. I really like your lucid explanation with examples, and willingness to share the code and data to go with it.
I have been exploring use of scispacy for EHR data, and was excited to find it had a NER component that can be normalized using UMLS. (From what I've learnt so far, you can use max_entities_per_mention and set it to 1 to get the highest scoring match. This also has the advantage of speeding up the parsing process compared to the default setting of 5.)
In my use case, that still leaves a *lot* of concepts to wade through. I'm looking at word/text embeddings (e.g. word2vec, text2vec) to help summarizing text without much loss of information.
Would appreciate any thoughts / pointers on this approach or alternatives. Thank you!
You're welcome Raj, and thanks for the kind words.
awesome .. thanks for sharing
A friend tried to run the code on an M1 mac and ran into some problems. He started with "a more recent version of blis" for older spaCy versions, but then said something about the adjacency matric being built by code not included.
Hi Jack, sorry for the delayed reply. I had stopped blogging for a bit and then I completely forgot about the comments, just noticed this. And you are right, my intent was to share the implementation of the algorithm (and the fact that it is effective) given the transition and emission matrices (which are created from the corpus ahead of time).
Hi Sujit, I tried to run your code with the given data but E_xindex.pkl is missing. Could you share the file or update the files in data section ?
Hi Anonymous, check out the [Data] link in the blog post, it points to a data repository where I provided the .pkl files. The E_xindex.pkl file is not provided explicitly but there is a notation under E_yindex.pkl that says "E_xindex.pkl is same as T_yindex.pkl (cp T_yindex.pkl E_xindex.pkl)".
Post a Comment