Last week a colleague and I were trying to figure out why his network would crash with a NaN (Not a Number) error some 20 or so epochs into training. Lately I have also become more interested in tuning neural networks, so this was a good opportunity for me to suggest fixes based on reasoning about the network. The network itself was built with Keras, like all the other networks our team has built from scratch so far, although we have adapted some third party networks written in Caffe and Tensorflow as well.
Now Keras is great for fast development because of its high level API. It results in very expressive code that reads like how you would actually visualize the network in your head or on a piece of paper. Also, because Keras automates away so many things and provides reasonable default values for many of its parameters, there are fewer things programmers can make mistakes about. For example, this awesome post on How to unit test machine learning code is based on Tensorflow, and while some of the cases mentioned are possible in Keras, they are much less likely.
However, while it is very easy to go from design to code in Keras, it is actually a little harder to work with, compared to say Tensorflow or Pytorch, when things go wrong and you have to figure out what. However, Keras does offer some tools and hooks that allow you to do this. In this post I talk about some of these that we (re-)discovered for ourselves last week. If you have favorites that I haven't included, please let me know in the comments.
The example I will use throughout this post is a simple fully connected network that I built to recognize MNIST images. The code to train and evailate this network can be found here. The code to define and compile it is as follows:
1 2 3 4 5 6 7 8 9 | model = Sequential()
model.add(Dense(512, activation="relu", input_shape=(784,)))
model.add(Dropout(0.2))
model.add(Dense(256, activation="relu"))
model.add(Dropout(0.2))
model.add(Dense(10, activation="softmax"))
model.compile(optimizer="adam", loss="categorical_crossentropy",
metrics=["accuracy"])
|
The first issue I have seen have have to do with sizing the intermediate tensors in the network. Keras only asks that you provide the dimensions of the input tensor(s), and it figure out the rest of the tensor dimensions automatically. The flip side of this convenience is that programmers may not realize what the dimensions are, and may make design errors based on this lack of understanding. Keras provides a model.summary() function that returns the output dimensions from each layer. I have found this very useful to get a better intuition about a network.
1 | model.summary()
|
Layer (type) Output Shape Param #
=================================================================
dense_4 (Dense) (None, 512) 401920
_________________________________________________________________
dropout_3 (Dropout) (None, 512) 0
_________________________________________________________________
dense_5 (Dense) (None, 256) 131328
_________________________________________________________________
dropout_4 (Dropout) (None, 256) 0
_________________________________________________________________
dense_6 (Dense) (None, 10) 2570
=================================================================
Total params: 535,818
Trainable params: 535,818
Non-trainable params: 0
_________________________________________________________________
If you need more granular information about the intermediate tensors, take a look at the About Keras layers page. You can get input shapes as well using some code like this:
1 2 | for layer in model.layers:
print(layer.name, layer.input.shape, layer.output.shape)
|
dense_1 (?, 784) (?, 512)
dropout_1 (?, 512) (?, 512)
dense_2 (?, 512) (?, 256)
dropout_2 (?, 256) (?, 256)
dense_3 (?, 256) (?, 10)
Another built-in diagnostic tool that I have been ignoring a bit so far is Tensorboard. Tensorboard was originally developed as part of the Tensorflow ecosystem, and allows Tensorflow developers to log certain things into a Tensorboard log file, which can later be used to visualize these logs graphically. The Keras project provides a way to write to Tensorboard using its TensorBoard callback. I learned to extract loss and other metrics from the output of model.fit() and plot it with matplotlib before the TensorBoard callback was popular, and have continued to use the approach mostly due to inertia. But the TensorBoard callback provides not only these plots, but the weight distributions for all the weights, biases and gradients. In case of networks where Embeddings and Images are involved, Tensorboard provides visualizations for them as well.
To invoke the Tensorboard callback, it needs to be defined and then declared in the callbacks queue in the model.fit() call.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 | tensorboard = TensorBoard(log_dir=TENSORBOARD_LOGS_DIR,
histogram_freq=1,
batch_size=BATCH_SIZE,
write_graph=True,
write_grads=True,
write_images=False,
embeddings_freq=0,
embeddings_layer_names=None,
embeddings_metadata=None)
...
history = model.fit(Xtrain, Ytrain, batch_size=BATCH_SIZE,
epochs=NUM_EPOCHS,
validation_split=0.1,
callbacks=[..., tensorboard, ...])
|
Here are the kind of visualizations you can expect on Tensorboard. The best resource I have found on interpreting these visualizations are Dandelion Mané's talk at Tensorflow Developers Summit 2017 and the Tensorboard documentation on Histograms
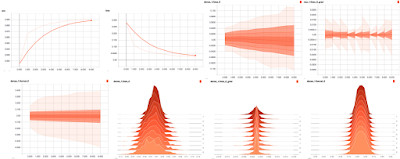
As nice as the Tensorboard callback is, it may not work for you all the time. For one thing, it appears that it doesn't work with fit_generator. You may also want to log values which are not meant to be logged with the Tensorboard callback. You can do that by writing your own callback in Keras.
Here is a callback that will capture the L2 norm, mean and standard deviation for each weight tensor in the network for each epoch and at the end of training, dump these values out to screen.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 | from keras import backend as K
from keras.callbacks import Callback
import numpy as np
def calc_stats(W):
return np.linalg.norm(W, 2), np.mean(W), np.std(W)
class MyDebugWeights(Callback):
def __init__(self):
super(MyDebugWeights, self).__init__()
self.weights = []
self.tf_session = K.get_session()
def on_epoch_end(self, epoch, logs=None):
for layer in self.model.layers:
name = layer.name
for i, w in enumerate(layer.weights):
w_value = w.eval(session=self.tf_session)
w_norm, w_mean, w_std = calc_stats(np.reshape(w_value, -1))
self.weights.append((epoch, "{:s}/W_{:d}".format(name, i),
w_norm, w_mean, w_std))
def on_train_end(self, logs=None):
for e, k, n, m, s in self.weights:
print("{:3d} {:20s} {:7.3f} {:7.3f} {:7.3f}".format(e, k, n, m, s))
|
The on_epoch_end and on_train_end are basically event handlers which fire off when the epoch has ended and when training has ended respectively. The Callback interface defines 6 such events, for the beginning and end of batch, epoch and training. See the Keras callbacks documentation for a list and some more examples.
You could use the callback above to train for a small number of epochs and observe how these attributes of the weight tensors change. At some point, I would like to write these values to disk and then read them and chart them maybe using something like Pandas, but my Pandas-fu is not strong enough for that at this time. Here is the output of after 2 wpochs of training.
Train on 54000 samples, validate on 6000 samples
Epoch 1/2
54000/54000 [==============================] - 4s - loss: 0.2830 - acc: 0.9146 - val_loss: 0.0979 - val_acc: 0.9718
Epoch 2/2
54000/54000 [==============================] - 3s - loss: 0.1118 - acc: 0.9663 - val_loss: 0.0758 - val_acc: 0.9773
0 dense_1/W_0 28.236 -0.002 0.045
0 dense_1/W_1 0.283 0.003 0.012
0 dense_2/W_0 20.631 0.002 0.057
0 dense_2/W_1 0.205 0.008 0.010
0 dense_3/W_0 4.962 -0.005 0.098
0 dense_3/W_1 0.023 -0.001 0.007
1 dense_1/W_0 30.455 -0.003 0.048
1 dense_1/W_1 0.358 0.003 0.016
1 dense_2/W_0 21.989 0.002 0.061
1 dense_2/W_1 0.273 0.010 0.014
1 dense_3/W_0 5.282 -0.008 0.104
1 dense_3/W_1 0.040 -0.002 0.013
Another thing we can do is to look at the attributes of the outputs at each layer. I initially tried to build this as another callback, but ran into some problems, then decided on this standalone implementation which can be called after every few epochs of training to see if anything has changed. This is adapted from the Keras FAQ.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 | def get_outputs(inputs, model):
layer_01_fn = K.function([model.layers[0].input, K.learning_phase()],
[model.layers[1].output])
layer_23_fn = K.function([model.layers[2].input, K.learning_phase()],
[model.layers[3].output])
layer_44_fn = K.function([model.layers[4].input, K.learning_phase()],
[model.layers[4].output])
layer_1_out = layer_01_fn([inputs, 1])[0]
layer_3_out = layer_23_fn([layer_1_out, 1])[0]
layer_4_out = layer_44_fn([layer_3_out, 1])[0]
return layer_1_out, layer_3_out, layer_4_out
out_1, out_3, out_4 = get_outputs(Xtest[0:10], model)
print("out_1", calc_stats(out_1))
print("out_3", calc_stats(out_3))
print("out_4", calc_stats(out_4))
|
I suspect we can make this more generic by looking up the model.layers data structure, but since it is kind of hard to forecast every kind of model you will build and because you will be doing this once per model, a quick and dirty implementation like the above may be preferable to something nicer. As before, we can rerun this every couple of epochs and get back the L2 norm, mean and standard deviation of the output tensors at each layer, as shown below.
out_1 (15.320195, 0.15846619, 0.36553052)
out_3 (31.983685, 0.52617866, 0.82984859)
out_4 (1.4138139, 0.1, 0.29160777)
Finally, we also wanted to figure out what the gradients looked like. The code for this adapted heavily from Edward Banner's comment in Keras Issue 2226. Like the code for visualizing the outputs, this code also needs to be run after training for a few epochs and compared with the previous values of L2 norm, mean and standard deviation for the gradients at different layers in the network.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 | def get_gradients(inputs, labels, model):
opt = model.optimizer
loss = model.total_loss
weights = model.weights
grads = opt.get_gradients(loss, weights)
grad_fn = K.function(inputs=[model.inputs[0],
model.sample_weights[0],
model.targets[0],
K.learning_phase()],
outputs=grads)
grad_values = grad_fn([inputs, np.ones(len(inputs)), labels, 1])
return grad_values
gradients = get_gradients(Xtest[0:10], Ytest[0:10], model)
for i in range(len(gradients)):
print("grad_{:d}".format(i), calc_stats(gradients[i]))
|
As before, the output below shows how the L2 norm, mean and standard deviation of the gradients at each layer. As with the output tensors, we train the network for 2 epochs, then then run this block of code. As you can guess, this sort of debugging works really well with an interactive development environment such as Jupyter Notebooks.
grad_0 (1.7725379, 1.1711028e-05, 0.0028093776)
grad_1 (0.17403033, 3.4195516e-05, 0.0076910509)
grad_2 (1.2508092, -7.3888972e-05, 0.003460743)
grad_3 (0.12154519, -0.00047613602, 0.0075816377)
grad_4 (1.5319482, 4.8748915e-11, 0.030318365)
grad_5 (0.10286356, -4.6566129e-11, 0.032528315)
That's all I had for today. The example network I have used here is quite simple, but these same ideas and tools can be used to debug more complex networks as well. These tools were built based on discussions betwwen my colleague and I last week, and the code is available here. I am sure many of you have your own favorite tools and tricks. If so, and you are okay with sharing, please let us know in the comments.













